🎆 ISMRM Raw Data¶
1️⃣ Shopping
👩🎤 NORAH JONES Readout¶
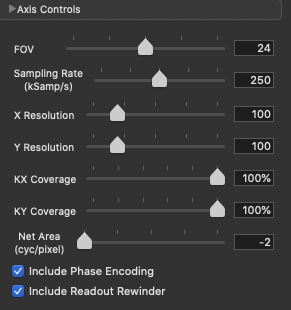
We are looking at the readout block of our NORAH JONES sequence /Sequence/CartesianReadout3D.spv. If you are an OSX user, you can inspect sequence components using SpinBench!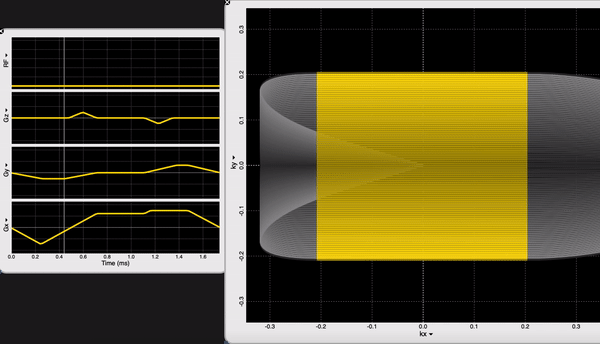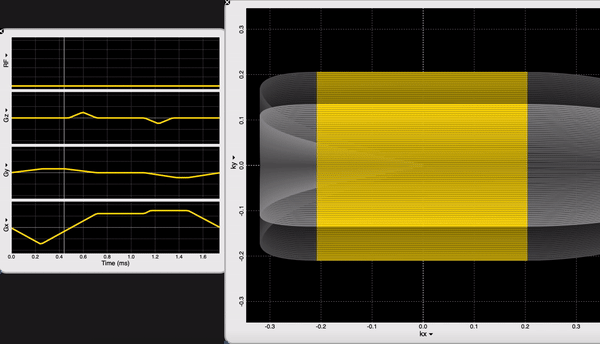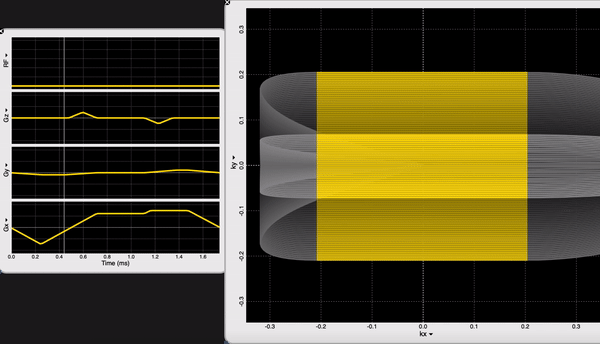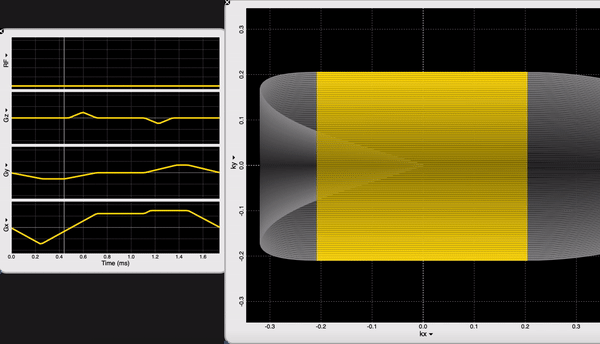
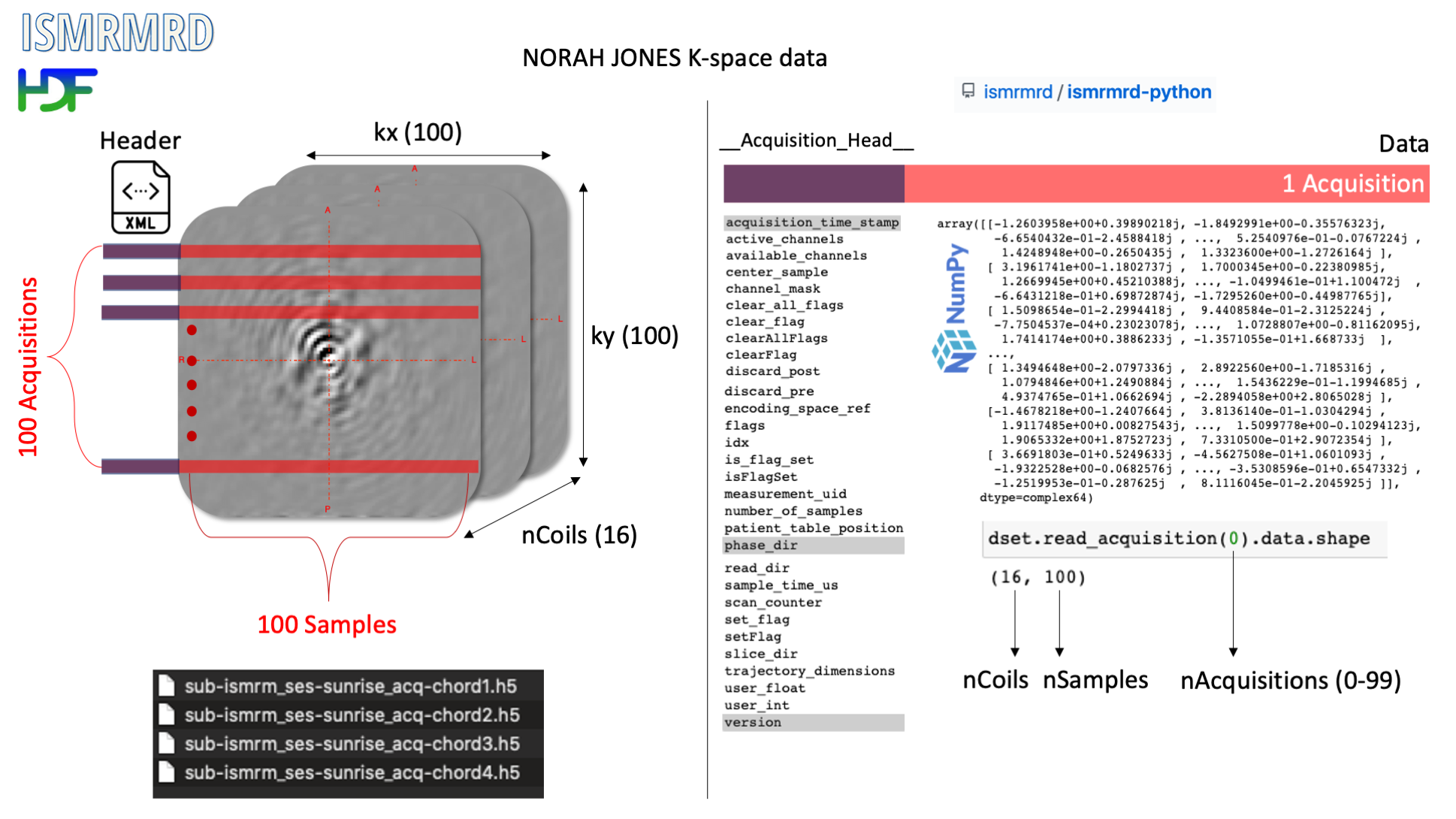
We have 100 acquisitions (or TRs or Phase Encodings) that are ordered outside-in to sample the center of the k-space the latest (steady-state). At each acquisition we have 100 samples. NORAH JONES sequence acquires all 4 images in this fashion, at the same location. You can see all the readout parameters below:
NORAH JONES k-space in ISMRM-RD¶
2️⃣ Mise en place
Let’s read and parse the data using NumPy relying purely on metadata provided by python-ismrmrd.
Import ISMRM-RD modules 🧲¶
from ismrmrd import Dataset as read_ismrmrd
from ismrmrd.xsd import CreateFromDocument as parse_ismrmd_header
import numpy as np
import plotly.express as px
import plotly.graph_objects as go
/Users/agah/opt/anaconda3/envs/jbnew/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
return f(*args, **kwds)
/Users/agah/opt/anaconda3/envs/jbnew/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning:
numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
/Users/agah/opt/anaconda3/envs/jbnew/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning:
numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
Create a simple list storing file names 🗂¶
files = list()
for chord in ['chord1','chord2','chord3','chord4']:
files.append('../KSpace/sub-ismrm_ses-sunrise_acq-' + chord + '.h5')
files
['../KSpace/sub-ismrm_ses-sunrise_acq-chord1.h5',
'../KSpace/sub-ismrm_ses-sunrise_acq-chord2.h5',
'../KSpace/sub-ismrm_ses-sunrise_acq-chord3.h5',
'../KSpace/sub-ismrm_ses-sunrise_acq-chord4.h5']
Explore acq-chord1 file 🕵️♀️¶
Read data & parse header¶
# Read the first file to explore data
dset = read_ismrmrd(files[0], 'dataset')
# Parse header
header = parse_ismrmd_header(dset.read_xml_header())
# See what's in the header
print('header has:',list(vars(header).keys()))
print('----')
print('header.encoding[0] has:',list(vars(header.encoding[0]).keys()))
header has: ['version', 'subjectInformation', 'studyInformation', 'measurementInformation', 'acquisitionSystemInformation', 'experimentalConditions', 'encoding', 'sequenceParameters', 'userParameters', 'waveformInformation']
----
header.encoding[0] has: ['encodedSpace', 'reconSpace', 'encodingLimits', 'trajectory', 'trajectoryDescription', 'parallelImaging', 'echoTrainLength']
Grab some useful metadata values¶
nX = header.encoding[0].encodedSpace.matrixSize.x
nY = header.encoding[0].encodedSpace.matrixSize.y
nZ = header.encoding[0].encodedSpace.matrixSize.z
nCoils = header.acquisitionSystemInformation.receiverChannels
print('nX: ', nX,'nY: ', nY,'nZ: ', nZ, 'nCoils: ', nCoils)
nX: 100 nY: 100 nZ: 1 nCoils: 16
Initialize a 3D NumPy matrix to store multi-channel raw data 🗃¶
We already know the dimensions! Given that we expect complex data, we can set dtype=complex64 😎 For simplicity I will omit nZ (we have one slice).
raw = np.zeros((nCoils, nX, nY), dtype=np.complex64)
print('Raw data shape: ', raw.shape)
Raw data shape: (16, 100, 100)
Loop over acquisitions to fill out raw¶
The dset object we created from the ISMRM-RD Dataset (we used alias read_ismrmrd) class has an important method: read_acquisition. We will loop over nY to read k-space stripes (cartesian) one by one.
for tr in range(nY):
raw[:,:,tr] = dset.read_acquisition(tr).data
Is not Plotly simply amazing?¶
This feature has been introduced in v4.14.0: with a single line of code you can animate or grid display 3D data.
fig = px.imshow(raw.real,animation_frame=0, color_continuous_scale='viridis', labels=dict(animation_frame="Channel"),template='plotly_dark')
fig.update_layout(title='Channel Raw')
fig
3️⃣ Time to perform a modest reconstruction :)
from scipy.fft import fft2, fftshift
from scipy import ndimage
Loop over channels and fft2¶
Tip: remove fftshift from fourier_ellipsoid function call, set filter size > 20, then play the animation along with your remix from the first notebook 🎵🎶
im = np.zeros(raw.shape)
# Let's apply some ellipsoid filter.
raw = ndimage.fourier_ellipsoid(fftshift(raw),size=2)
#raw = ndimage.fourier_ellipsoid(raw,size=2)
for ch in range(nCoils):
# Comment in and see what it gives
im[ch,:,:] = abs(fftshift(fft2(raw[ch,:,:])))
# Normalize
im[ch,:,:] /= im[ch,:,:].max()
fig = px.imshow(im,color_continuous_scale='viridis', animation_frame=0,template='plotly_dark')
fig.update_layout(title='Channel Recon').show()
fig
im_sos = np.sqrt(np.sum(im*np.conj(im),axis=0))
# Normalize
im_sos /= im_sos.max()
# Crop a bit
fig = px.imshow(im_sos[10:90,0:80],template='plotly_dark',color_continuous_scale='viridis')
fig.update_layout(title='Sum of Squares Combine')
fig
Exercises
Rotate/flip images
Reconstruct
SoSimages for every TR by writing some functions and collect them into a 3D matrix.Display voxel intensity changes over TRs at the center of each (14) sphere